Brain Tumor Segmentation (BraTS) Challenge 2021 Homepage
github项目地址 brats-unet: UNet for brain tumor segmentation
BraTS是MICCAI所有比赛中历史最悠久的,到2021年已经连续举办了10年,参赛人数众多,是学习医学图像分割最前沿的平台之一。
1.数据准备
简介:
比赛方提供多机构、多参数多模态核磁共振成像(mpMRI)数据集,包括训练集(1251例)和验证集(219例)以及测试集(530例),一共2000例患者的mpMRI扫描结果。其中训练集包含图像和分割标签,验证集和测试集没有分割标签,验证集被用于公共排行榜,测试集不公开,用作参赛者的最终排名评测。
四种模态数据:flair, t1ce, t1, t2,每个模态的数据大小都为 240 x 240 x 155,且共享分割标签。
分割标签:[0, 1, 2, 4]
- label0:背景(background)
- label1:坏疽(NT, necrotic tumor core)
- label2:浮肿区域(ED,peritumoral edema)
- label4:增强肿瘤区域(ET,enhancing tumor)
本次比赛包括两个任务:
- Task1:mpMRI扫描中分割内在异质性脑胶质母细胞瘤区域
- Task2:预测术前基线扫描中的MGMT启动子甲基化状态
本文从数据处理、评价指标、损失函数、模型训练四个方面介绍Task1的整体实现过程
数据集下载地址:
1.官网:BraTS 2021 Challenge 需要注册和申请(包括训练集和验证集)
2.Kaggle:BRaTS 2021 Task 1 Dataset 建议在kaggle上下载,数据集与官网一致(不包括验证集)
数据准备:
下载数据集,解压后如下图所示:
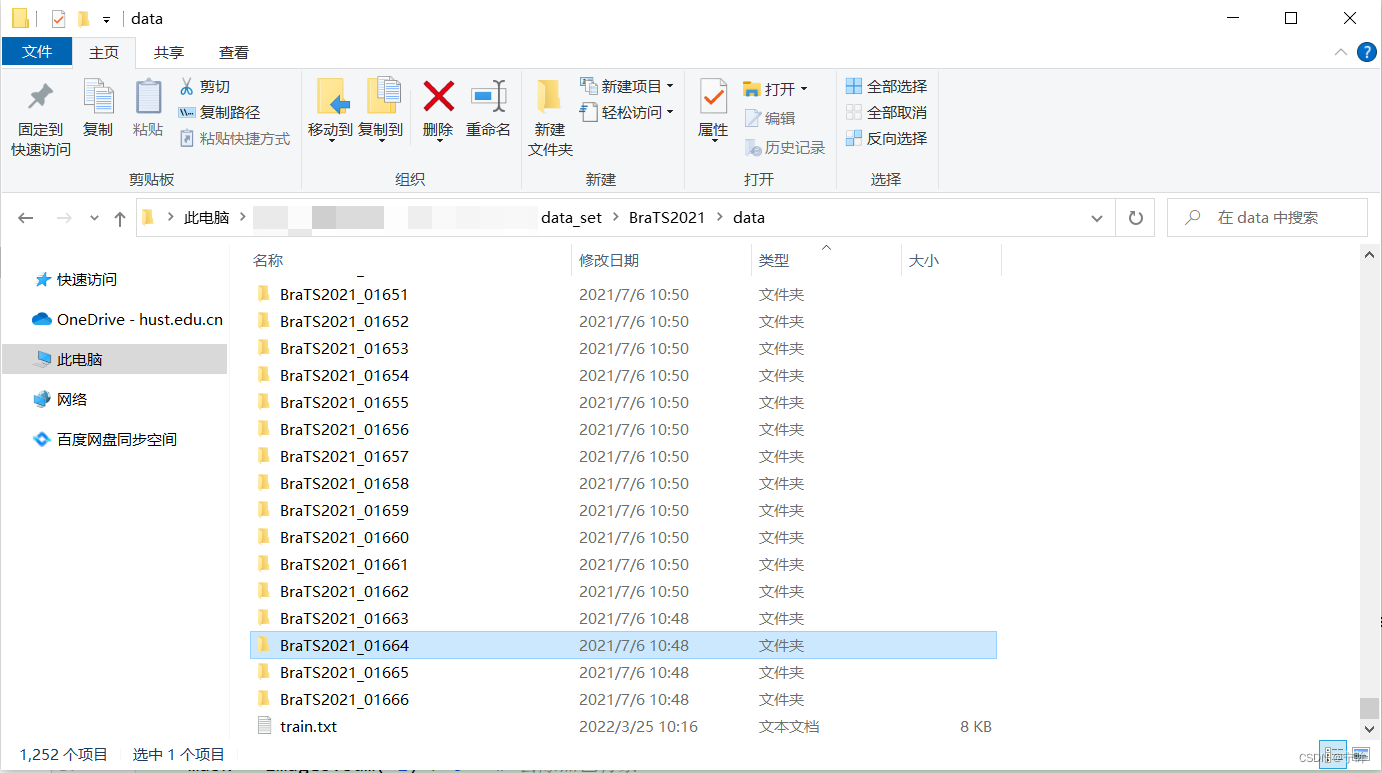
每个病例包含四种模态的MRI图像和分割标签,结构如下:
BraTS2021_00000 ├── BraTS2021_00000_flair.nii.gz ├── BraTS2021_00000_seg.nii.gz ├── BraTS2021_00000_t1ce.nii.gz ├── BraTS2021_00000_t1.nii.gz └── BraTS2021_00000_t2.nii.gz
建议使用3D Slicer查看图像和标签,直观的了解一下自己要用的数据集。
2.数据预处理
每个病例的四种MRI图像大小为 240 x 240 x 155,且共享标签。
鉴于此,我将四种模态的图像合并为一个4D图像(C x H x W x D , C=4),并且和分割标签一起保存为一个.h5文件,方便后续处理。
import h5py import os import numpy as np import SimpleITK as sitk from tqdm import tqdm # 四种模态的mri图像 modalities = ('flair', 't1ce', 't1', 't2') # train train_set = { 'root': '/data/omnisky/postgraduate/Yb/data_set/BraTS2021/data', # 四个模态数据所在地址 'out': '/data/omnisky/postgraduate/Yb/data_set/BraTS2021/dataset/', # 预处理输出地址 'flist': 'train.txt', # 训练集名单(有标签) }- 将图像保存为32位浮点数(np.float32),标签保存为整数(np.uint8),写入.h5文件
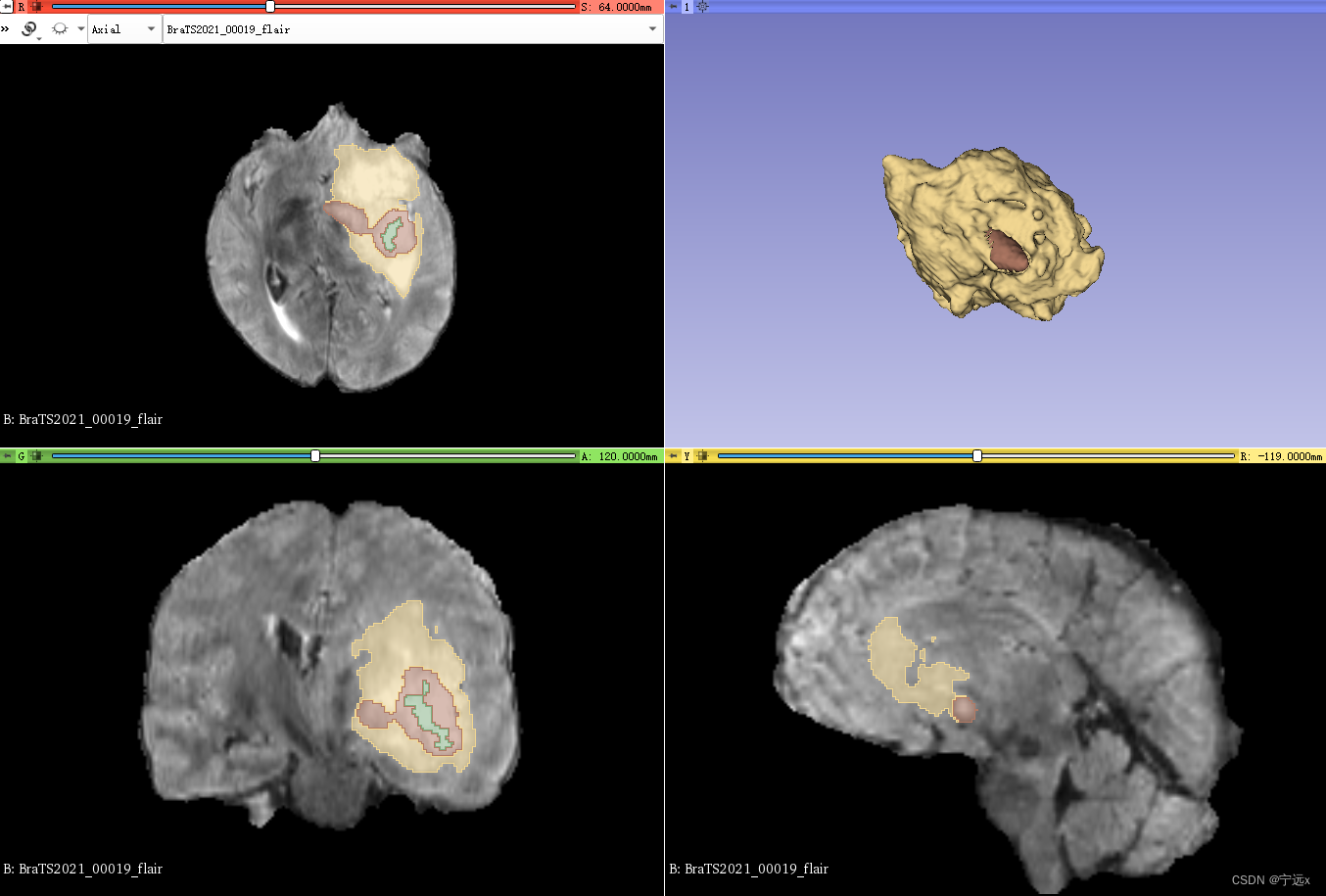
- 对每张图像的灰度进行标准化,但保持背景区域为0
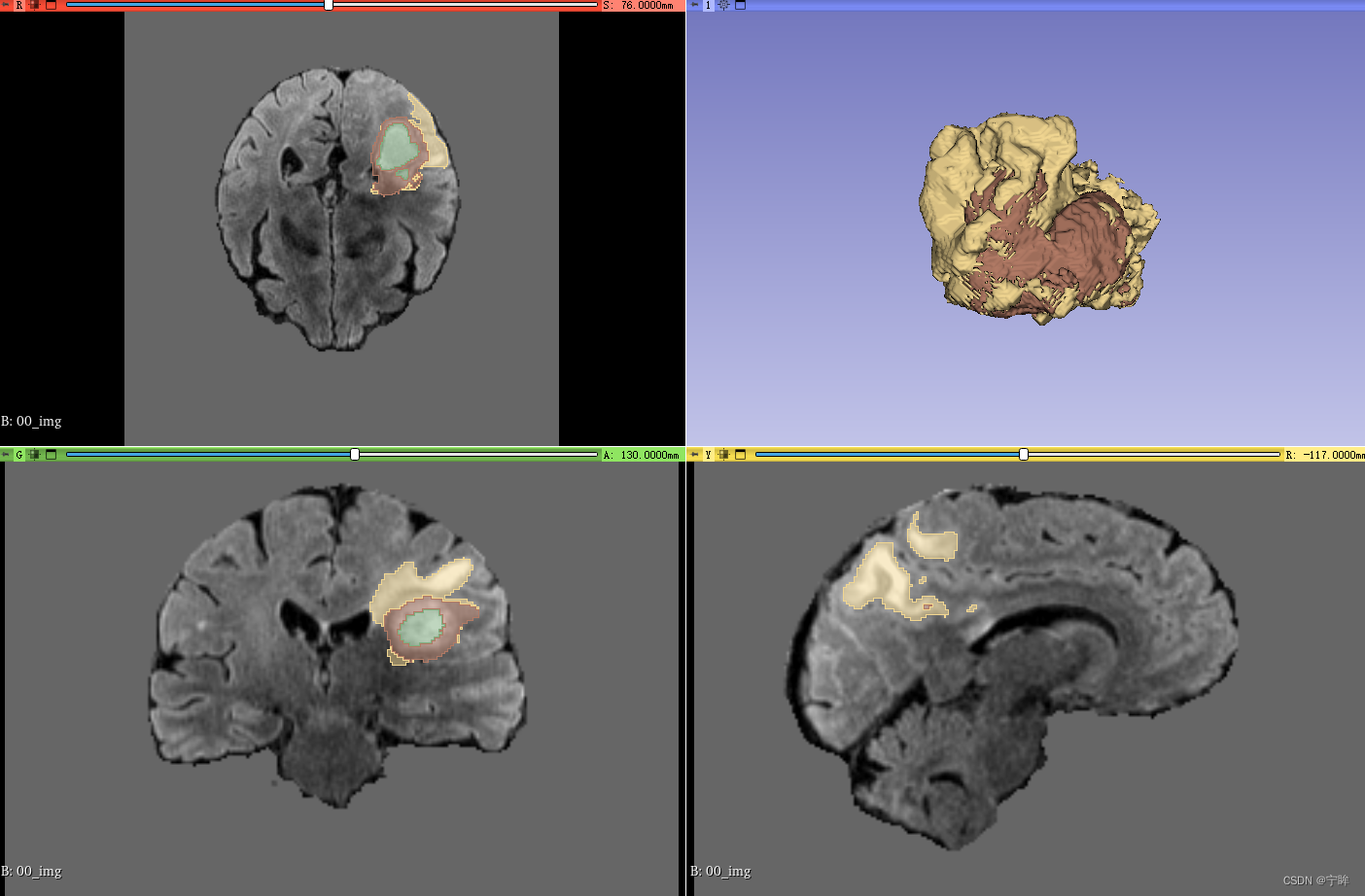
- 上图是预处理后的图像,背景区域为0
def process_h5(path, out_path): """ Save the data with dtype=float32. z-score is used but keep the background with zero! """ # SimpleITK读取图像默认是是 DxHxW,这里转为 HxWxD label = sitk.GetArrayFromImage(sitk.ReadImage(path + 'seg.nii.gz')).transpose(1,2,0) print(label.shape) # 堆叠四种模态的图像,4 x (H,W,D) -> (4,H,W,D) images = np.stack([sitk.GetArrayFromImage(sitk.ReadImage(path + modal + '.nii.gz')).transpose(1,2,0) for modal in modalities], 0) # [240,240,155] # 数据类型转换 label = label.astype(np.uint8) images = images.astype(np.float32) case_name = path.split('/')[-1] # case_name = os.path.split(path)[-1] # Windows路径与linux不同 path = os.path.join(out_path,case_name) output = path + 'mri_norm2.h5' # 对第一个通道求和,如果四个模态都为0,则标记为背景(False) mask = images.sum(0) > 0 for k in range(4): x = images[k,...] # y = x[mask] # 对背景外的区域进行归一化 x[mask] -= y.mean() x[mask] /= y.std() images[k,...] = x print(case_name,images.shape,label.shape) f = h5py.File(output, 'w') f.create_dataset('image', data=images, compression="gzip") f.create_dataset('label', data=label, compression="gzip") f.close() def doit(dset): root, out_path = dset['root'], dset['out'] file_list = os.path.join(root, dset['flist']) subjects = open(file_list).read().splitlines() names = ['BraTS2021_' + sub for sub in subjects] paths = [os.path.join(root, name, name + '_') for name in names] for path in tqdm(paths): process_h5(path, out_path) # break print('Finished') if __name__ == '__main__': doit(train_set)数据保存在 mri_norm2.h5 文件中,每个 mri_norm2.h5 相当于一个字典,字典的键为 image 和 label ,值为对应的数组。
处理后的数据,可以用下面的几行代码测试一下,记得修改为你自己的路径
import h5py import numpy as np p = '/***/data_set/BraTS2021/all/BraTS2021_00000_mri_norm2.h5' h5f = h5py.File(p, 'r') image = h5f['image'][:] label = h5f['label'][:] print('image shape:',image.shape,'\t','label shape',label.shape) print('label set:',np.unique(label)) # image shape: (4, 240, 240, 155) label shape (240, 240, 155) # label set: [0 1 2 4]将数据集按照 8:1:1随机划分为训练集、验证集和测试集,将划分后的数据名保存为.txt文件
import os from sklearn.model_selection import train_test_split # 预处理输出地址 data_path = "/***/data_set/BraTS2021/dataset" train_and_test_ids = os.listdir(data_path) train_ids, val_test_ids = train_test_split(train_and_test_ids, test_size=0.2,random_state=21) val_ids, test_ids = train_test_split(val_test_ids, test_size=0.5,random_state=21) print("Using {} images for training, {} images for validation, {} images for testing.".format(len(train_ids),len(val_ids),len(test_ids))) with open('/***/data_set/BraTS2021/train.txt','w') as f: f.write('\n'.join(train_ids)) with open('/***/data_set/BraTS2021/valid.txt','w') as f: f.write('\n'.join(val_ids)) with open('/***/data_set/BraTS2021/test.txt','w') as f: f.write('\n'.join(test_ids))划分结果:
Using 1000 images for training, 125 images for validation, 126 images for testing. ...... BraTS2021_00002_mri_norm2.h5 BraTS2021_00003_mri_norm2.h5 BraTS2021_00014_mri_norm2.h5 ......
3.数据增强
下面是我写的Dataset类以及一些数据增强方法
整体架构
import os import torch from torch.utils.data import Dataset import random import numpy as np from torchvision.transforms import transforms import h5py class BraTS(Dataset): def __init__(self,data_path, file_path,transform=None): with open(file_path, 'r') as f: self.paths = [os.path.join(data_path, x.strip()) for x in f.readlines()] self.transform = transform def __getitem__(self, item): h5f = h5py.File(self.paths[item], 'r') image = h5f['image'][:] label = h5f['label'][:] #[0,1,2,4] -> [0,1,2,3] label[label == 4] = 3 # print(image.shape) sample = {'image': image, 'label': label} if self.transform: sample = self.transform(sample) return sample['image'], sample['label'] def __len__(self): return len(self.paths) def collate(self, batch): return [torch.cat(v) for v in zip(*batch)] if __name__ == '__main__': from torchvision import transforms data_path = "/***/data_set/BraTS2021/dataset" test_txt = "/***/data_set/BraTS2021/test.txt" test_set = BraTS(data_path,test_txt,transform=transforms.Compose([ RandomRotFlip(), RandomCrop((160,160,128)), GaussianNoise(p=0.1), ToTensor() ])) d1 = test_set[0] image,label = d1 print(image.shape) print(label.shape) print(np.unique(label))具体的数据增强方法我列在了下面,包括裁剪、旋转、翻转、高斯噪声、对比度变换和亮度增强的源码,部分代码借鉴了nnUNet的数据增强方法。
随机裁剪
原始图像尺寸为 240 x 240 x 155,但图像周围是有很多黑边的,我将图像裁剪为 160 x 160 x 128
class RandomCrop(object): """ Crop randomly the image in a sample Args: output_size (int): Desired output size """ def __init__(self, output_size): self.output_size = output_size def __call__(self, sample): image, label = sample['image'], sample['label'] (c, w, h, d) = image.shape w1 = np.random.randint(0, w - self.output_size[0]) h1 = np.random.randint(0, h - self.output_size[1]) d1 = np.random.randint(0, d - self.output_size[2]) label = label[w1:w1 + self.output_size[0], h1:h1 + self.output_size[1], d1:d1 + self.output_size[2]] image = image[:,w1:w1 + self.output_size[0], h1:h1 + self.output_size[1], d1:d1 + self.output_size[2]] return {'image': image, 'label': label}中心裁剪
class CenterCrop(object): def __init__(self, output_size): self.output_size = output_size def __call__(self, sample): image, label = sample['image'], sample['label'] (c,w, h, d) = image.shape w1 = int(round((w - self.output_size[0]) / 2.)) h1 = int(round((h - self.output_size[1]) / 2.)) d1 = int(round((d - self.output_size[2]) / 2.)) label = label[w1:w1 + self.output_size[0], h1:h1 + self.output_size[1], d1:d1 + self.output_size[2]] image = image[:,w1:w1 + self.output_size[0], h1:h1 + self.output_size[1], d1:d1 + self.output_size[2]] return {'image': image, 'label': label}随机翻转
旋转可能会导致图像重采样,因为数据集比较充分,我只在{90,180,270}度做一个简单旋转,不涉及重采样。
class RandomRotFlip(object): """ Crop randomly flip the dataset in a sample Args: output_size (int): Desired output size """ def __call__(self, sample): image, label = sample['image'], sample['label'] k = np.random.randint(0, 4) image = np.stack([np.rot90(x,k) for x in image],axis=0) label = np.rot90(label, k) axis = np.random.randint(1, 4) image = np.flip(image, axis=axis).copy() label = np.flip(label, axis=axis-1).copy() return {'image': image, 'label': label}高斯噪声
def augment_gaussian_noise(data_sample, noise_variance=(0, 0.1)): if noise_variance[0] == noise_variance[1]: variance = noise_variance[0] else: variance = random.uniform(noise_variance[0], noise_variance[1]) data_sample = data_sample + np.random.normal(0.0, variance, size=data_sample.shape) return data_sample class GaussianNoise(object): def __init__(self, noise_variance=(0, 0.1), p=0.5): self.prob = p self.noise_variance = noise_variance def __call__(self, sample): image = sample['image'] label = sample['label'] if np.random.uniform()对比度变换
- contrast_range:对比度增强的范围
- preserve_range:是否保留数据的取值范围
- per_channel:是否对每个通道的图像分别进行对比度增强
def augment_contrast(data_sample, contrast_range=(0.75, 1.25), preserve_range=True, per_channel=True): if not per_channel: mn = data_sample.mean() if preserve_range: minm = data_sample.min() maxm = data_sample.max() if np.random.random() maxm] = maxm else: for c in range(data_sample.shape[0]): mn = data_sample[c].mean() if preserve_range: minm = data_sample[c].min() maxm = data_sample[c].max() if np.random.random() maxm] = maxm return data_sample class ContrastAugmentationTransform(object): def __init__(self, contrast_range=(0.75, 1.25), preserve_range=True, per_channel=True,p_per_sample=1.): self.p_per_sample = p_per_sample self.contrast_range = contrast_range self.preserve_range = preserve_range self.per_channel = per_channel def __call__(self, sample): image = sample['image'] label = sample['label'] for b in range(len(image)): if np.random.uniform()亮度变换
附加亮度从具有μ和σ的高斯分布中采样
def augment_brightness_additive(data_sample, mu:float, sigma:float , per_channel:bool=True, p_per_channel:float=1.): if not per_channel: rnd_nb = np.random.normal(mu, sigma) for c in range(data_sample.shape[0]): if np.random.uniform() 'image': data, 'label': label} 'image': image, 'label': label} 'loss':loss,'dice1':dice1,'dice2':dice2,'dice3':dice3} def val_loop(model,criterion,val_loader,device): model.eval() running_loss = 0 dice1_val = 0 dice2_val = 0 dice3_val = 0 pbar = tqdm(val_loader) with torch.no_grad(): for images, masks in pbar: images, masks = images.to(device), masks.to(device) outputs = model(images) # outputs = torch.softmax(outputs,dim=1) loss = criterion(outputs, masks) dice1, dice2, dice3 = cal_dice(outputs, masks) running_loss += loss.item() dice1_val += dice1.item() dice2_val += dice2.item() dice3_val += dice3.item() # pbar.desc = "loss:{:.3f} dice1:{:.3f} dice2:{:.3f} dice3:{:.3f} ".format(loss,dice1,dice2,dice3) loss = running_loss / len(val_loader) dice1 = dice1_val / len(val_loader) dice2 = dice2_val / len(val_loader) dice3 = dice3_val / len(val_loader) return {'loss':loss,'dice1':dice1,'dice2':dice2,'dice3':dice3} def train(model,optimizer,scheduler,criterion,train_loader, val_loader,epochs,device,train_log,valid_loss_min=999.0): for e in range(epochs): # train for epoch train_metrics = train_loop(model,optimizer,scheduler,criterion,train_loader,device,e) # eval for epoch val_metrics = val_loop(model,criterion,val_loader,device) info1 = "Epoch:[{}/{}] train_loss: {:.3f} valid_loss: {:.3f} ".format(e+1,epochs,train_metrics["loss"],val_metrics["loss"]) info2 = "Train--ET: {:.3f} TC: {:.3f} WT: {:.3f} ".format(train_metrics['dice1'],train_metrics['dice2'],train_metrics['dice3']) info3 = "Valid--ET: {:.3f} TC: {:.3f} WT: {:.3f} ".format(val_metrics['dice1'],val_metrics['dice2'],val_metrics['dice3']) print(info1) print(info2) print(info3) with open(train_log,'a') as f: f.write(info1 + '\n' + info2 + ' ' + info3 + '\n') if not os.path.exists(args.save_path): os.makedirs(args.save_path) save_file = {"model": model.state_dict(), "optimizer": optimizer.state_dict()} if val_metrics['loss']
- 上图是预处理后的图像,背景区域为0